The main goal of mpathr is to provide functions to
import data from the m-Path platform, as well as provide functions for
common manipulations for ESM data.
Importing m-Path data
To show how to import data using mpathr, we provide
example data within the package:
mpath_example()
#> [1] "example_basic.csv" "example_meta.csv"As shown above, the package comes with an example of the
basic.csv that can be exported from the m-Path
platform.
To read this data into R, we can use the read_mpath()
function. We will also need a path to the meta data. The meta data is a
file that contains information about the data types of each column, as
well as the possible responses for categorical columns.
The main advantage of using read_mpath(), as opposed to
other functions like read.csv(), is that
read_mpath() uses the meta data to correctly interpret the
data types. Furthermore it will also automatically convert columns that
store multiple responses into lists. For a response with multiple
options like 1,4,6, read_mpath() will store a
list with each number, which facilitates further preprocessing of these
responses.
We can obtain the paths to the example basic data and meta data using
the mpath_example() function:
# find paths to example basic and meta data:
basic_path <- mpath_example(file = "example_basic.csv")
meta_path <- mpath_example("example_meta.csv")
# read the data
data <- read_mpath(
file = basic_path,
meta_data = meta_path
)
data
#> # A tibble: 2,221 × 100
#> connectionId legacyCode code alias initials accountCode scheduledBeepId
#> <int> <chr> <chr> <chr> <chr> <chr> <int>
#> 1 234609 !9v48@jp7a7 !byyo kj… abc Ver jp7a7 -1
#> 2 234609 !9v48@jp7a7 !byyo kj… abc Ver jp7a7 28626776
#> 3 234609 !9v48@jp7a7 !byyo kj… abc Ver jp7a7 28626777
#> 4 234609 !9v48@jp7a7 !byyo kj… abc Ver jp7a7 28626781
#> 5 234609 !9v48@jp7a7 !byyo kj… abc Ver jp7a7 28626782
#> 6 234609 !9v48@jp7a7 !byyo kj… abc Ver jp7a7 28626784
#> 7 234609 !9v48@jp7a7 !byyo kj… abc Ver jp7a7 28626785
#> 8 234609 !9v48@jp7a7 !byyo kj… abc Ver jp7a7 28626786
#> 9 234609 !9v48@jp7a7 !byyo kj… abc Ver jp7a7 28626796
#> 10 234609 !9v48@jp7a7 !byyo kj… abc Ver jp7a7 28626795
#> # ℹ 2,211 more rows
#> # ℹ 93 more variables: sentBeepId <int>, reminderForOriginalSentBeepId <int>,
#> # questionListName <chr>, questionListLabel <chr>, fromProtocolName <chr>,
#> # timeStampScheduled <int>, timeStampSent <int>, timeStampStart <int>,
#> # timeStampStop <int>, originalTimeStampSent <int>, timeZoneOffset <int>,
#> # deltaUTC <dbl>, consent_yesno_yesno <int>,
#> # gender_multipleChoice_index <int>, gender_multipleChoice_string <chr>, …Saving m-Path data
The resulting data frame will contain columns with lists, which can be problematic when saving the data. To save the data, we suggest the following two options:
If you want to save the data as a comma-separated values (CSV) file
to use it in another program, use write_mpath(). This
function will collapse most list columns to a single string and parses
all character columns to JSON strings, essentially reversing the
operations performed by read_mpath(). Note that this does
not mean that data can be read back using read_mpath(),
because the data may have been modified and thus no longer be in line
with the meta data.
write_mpath(
x = data,
file = "data.csv"
)Otherwise, if the data will be used exclusively in R, we suggest saving it as an R object (.RData or .RDS):
Obtaining response rates
response_rate function
Some common operations that are done on Experience Sampling
Methodology (ESM) data have to do with the participants’ response rate.
We provide a function response_rate() that calculates the
response_rate per participant for the entire duration of the study, or
for a specific time frame.
This function takes as argument a valid_col, that takes
a logical column that stores whether the beep was answered by the
participant, or not, as well as a participant_col, that
identifies each distinct participant.
We will show how to use this function with the
example_data, that contains data from the same study as the
example_basic.csv file, but after some cleaning.
example_data
#> # A tibble: 1,980 × 47
#> participant code questionnaire scheduled sent
#> <int> <chr> <chr> <dttm> <dttm>
#> 1 2 !bxxm dqfu main_question… 2024-04-24 08:00:57 2024-04-24 08:00:59
#> 2 2 !bxxm dqfu main_question… 2024-04-24 09:25:44 2024-04-24 09:25:45
#> 3 2 !bxxm dqfu main_question… 2024-04-24 11:14:18 2024-04-24 11:14:20
#> 4 2 !bxxm dqfu main_question… 2024-04-24 12:58:05 2024-04-24 12:58:06
#> 5 2 !bxxm dqfu main_question… 2024-04-24 14:19:51 2024-04-24 14:19:52
#> 6 2 !bxxm dqfu main_question… 2024-04-24 15:43:05 2024-04-24 15:43:06
#> 7 2 !bxxm dqfu main_question… 2024-04-24 17:12:03 2024-04-24 17:12:04
#> 8 2 !bxxm dqfu main_question… 2024-04-24 18:07:23 2024-04-24 18:07:25
#> 9 2 !bxxm dqfu main_question… 2024-04-24 20:01:21 2024-04-24 20:01:22
#> 10 2 !bxxm dqfu main_question… 2024-04-24 21:00:14 2024-04-24 21:00:17
#> # ℹ 1,970 more rows
#> # ℹ 42 more variables: start <dttm>, stop <dttm>, phone_server_offset <dbl>,
#> # obs_n <int>, day_n <int>, obs_n_day <int>, answered <lgl>, bpm_day <dbl>,
#> # gender <int>, gender_string <chr>, age <chr>, life_satisfaction <dbl>,
#> # neuroticism <dbl>, slider_happy <int>, slider_sad <int>,
#> # slider_angry <int>, slider_relaxed <int>, slider_anxious <int>,
#> # slider_energetic <int>, slider_tired <int>, location_index <int>, …
response_rates <- response_rate(
data = example_data,
valid_col = answered,
participant_col = participant
)
#> Calculating response rates for the entire duration of the study.
response_rates
#> # A tibble: 18 × 3
#> participant number_of_beeps response_rate
#> <int> <int> <dbl>
#> 1 2 110 0.418
#> 2 3 110 0.564
#> 3 4 110 0.845
#> 4 5 110 0.9
#> 5 6 110 0.664
#> 6 7 110 0.673
#> 7 9 110 0.545
#> 8 10 110 0.873
#> 9 11 110 0.836
#> 10 12 110 0.9
#> 11 13 110 0.8
#> 12 14 110 0.755
#> 13 15 110 0.682
#> 14 16 110 0.318
#> 15 17 110 0.791
#> 16 18 110 0.818
#> 17 19 110 0.636
#> 18 20 110 0.436The function returns a data frame with:
- The
participantcolumn, as specified inparticipant_col - The
number_of_beepsused to calculate the response rate. - The
response_ratecolumn, which is the proportion of valid responses (specified invalid_col) per participant.
The output of this function can further be used to identify participants with low response rates:
response_rates[response_rates$response_rate < 0.5,]
#> # A tibble: 3 × 3
#> participant number_of_beeps response_rate
#> <int> <int> <dbl>
#> 1 2 110 0.418
#> 2 16 110 0.318
#> 3 20 110 0.436We could also be interested in seeing the participants’ response rate
during a specific period of time (for example, if we think a
participant’s compliance significantly dropped a certain date). In this
case, we should supply the function with the (otherwise optional)
argument time_col, that should contain times stored as
POSIXct objects, and specify the date period that we are
interested in (in the format yyyy-mm-dd or
yyyy/mm/dd):
response_rates_after_15 <- response_rate(
data = example_data,
valid_col = answered,
participant_col = participant,
time_col = sent,
period_start = '2024-05-15'
)
#> Calculating response rates starting from date: 2024-05-15This will return the participant’s response rate after the 15th of May 2024.
response_rates_after_15
#> # A tibble: 5 × 3
#> participant number_of_beeps response_rate
#> <int> <int> <dbl>
#> 1 16 55 0.0364
#> 2 17 55 0.691
#> 3 18 110 0.818
#> 4 19 110 0.636
#> 5 20 110 0.436plot_response_rate function
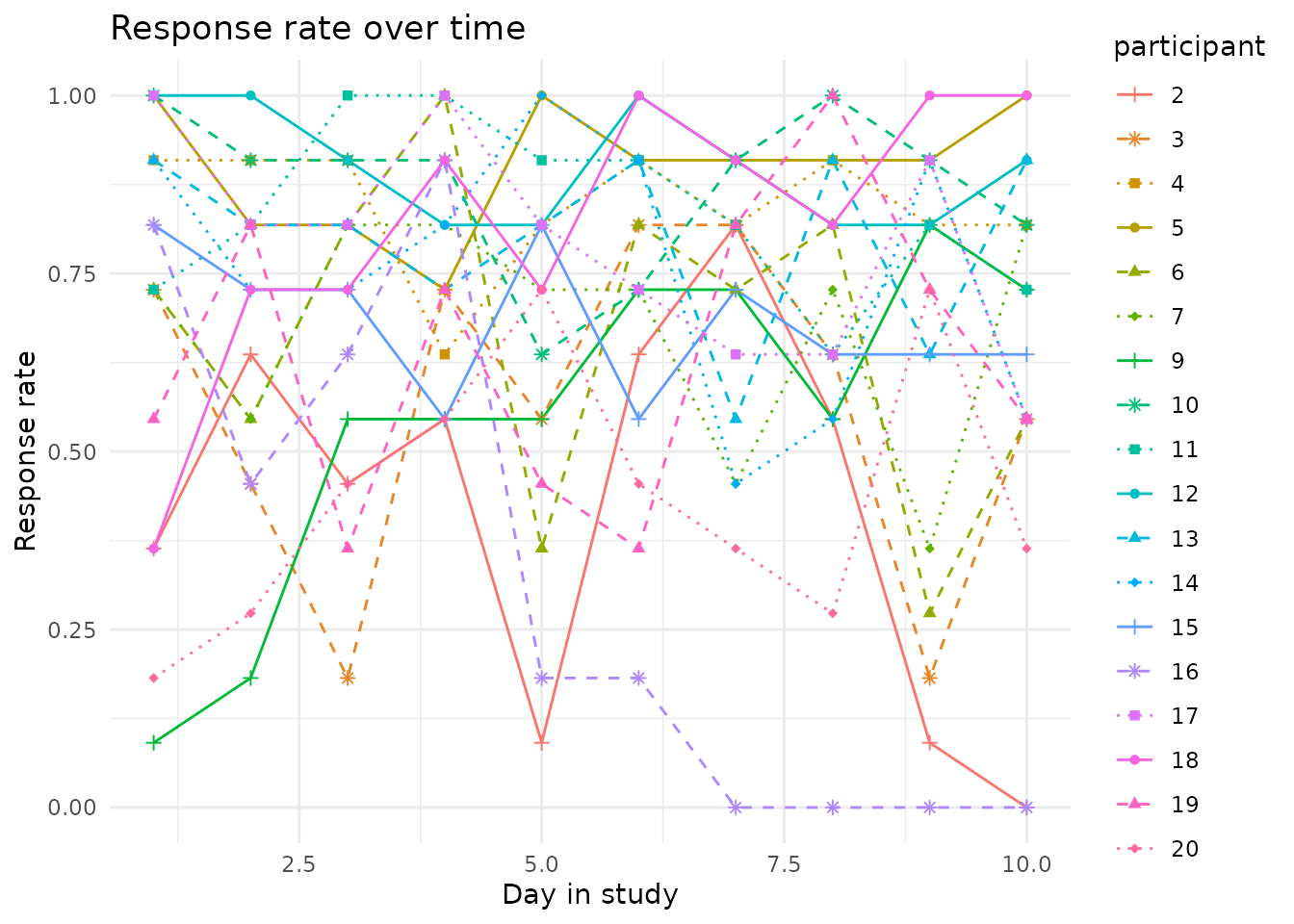
We also suggest a way to plot the participant response rates, to
identify patterns like response rates dropping over time. For this, we
provide the plot_response_rate() function.
plot_response_rate(
data = example_data,
time_col = sent,
participant_col = participant,
valid_col = answered
)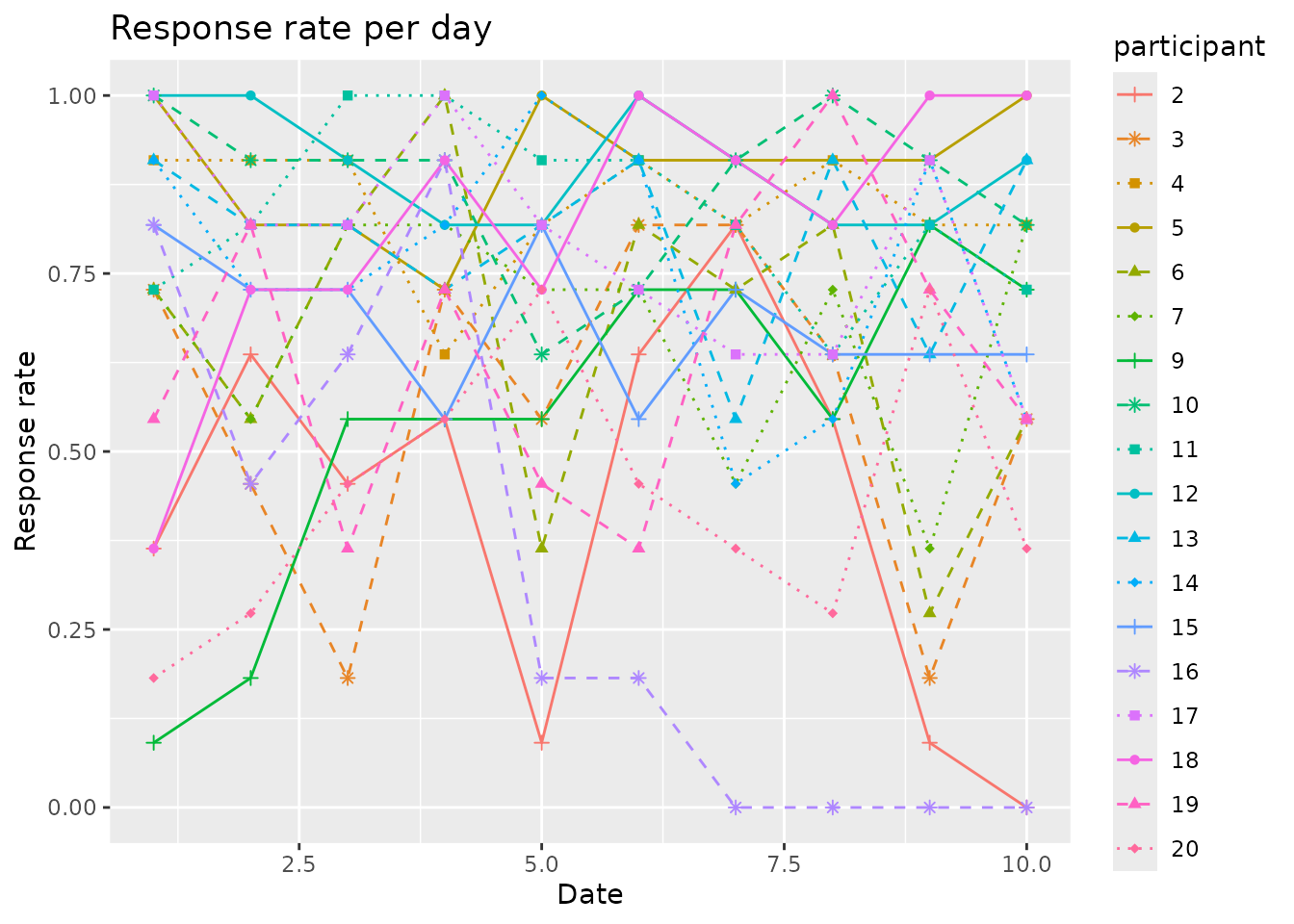 Note that the resulting plot can be further customized using the
Note that the resulting plot can be further customized using the
ggplot2 package.
library(ggplot2)
plot_response_rate(
data = example_data,
time_col = sent,
participant_col = participant,
valid_col = answered
) +
theme_minimal() +
ggtitle('Response rate over time') +
xlab('Day in study')